
For the last two and a half million years or so — up until a certain species of upright-walking ape descendants really started making their presence known — the greatest force shaping Earth’s biological diversity may well have been ice. I’m talking, of course, about the global glacial cycles of the Pleistocene, a geological period during which the planet’s climate cycled between warmer periods and cooler ages in which the polar ice sheets advanced towards the equator.
These cycles left their mark on the world we see today, especially in the distributions of species that survived through the most recent cycle of glacial advance and retreat. Species that lived in temperate latitudes would have migrated (if they could) equator-ward as the ice caps expanded, and back towards the poles as they melted again. That migration remixed ecological communities and individual species everywhere the glaciers touched. It would have left a straightforward signature in the genetic diversity of any species that experienced it — and as an impressive analysis published earlier this summer in Evolution Letters shows, that signature is a global latitudinal gradient of genetic diversity.
On the one hand, losing habitat to advancing glaciers would mean that many species in temperate latitudes lost population size when the climate cooled — and thereby diversity, because smaller populations are more prone to lose diversity via genetic drift. On the other hand, an expanding population — such as a species that is gaining new habitat as glaciers retreat — is more likely to have lots of rare genetic variants, carried by just one or a few individuals. This is because, while the average individual in the population is producing more offspring than necessary to maintain the population size, mutation introduces new variants faster than drift removes them.
The new study, led by Emanuel Fonseca, demonstrates this pattern in tens of thousands of species represented in public databases of DNA sequence data. The coauthors used a utility called phylogatR to aggregate mitochondrial and chloroplast sequence data and the associated geographic locations of sample collection in thousands of published studies. They dropped datasets with insufficient sampling, leaving 38,134 species of amphibians, birds, fish, mammals, reptiles, insects, arachnids, and plants. For each species-location dataset, they calculated the standard nucleotide diversity statistic π, the average number of site differences between pairs of sequences, as well as Tajima’s D and R2, which measure patterns in sequence diversity that reflect changes in population size. (Mitochondrial and chloroplast sequences have two useful features: they’ve been used in studies going back decades, and they have the same effective ploidy across species, so they don’t have to account for polyploidy in the calculation.)
The core analysis, estimated nucleotide diversity, is presented in a figure you can expect to find in evolutionary biology textbooks for years to come, as well as the slides I’ll use in my lecture on genetic drift this fall semester.
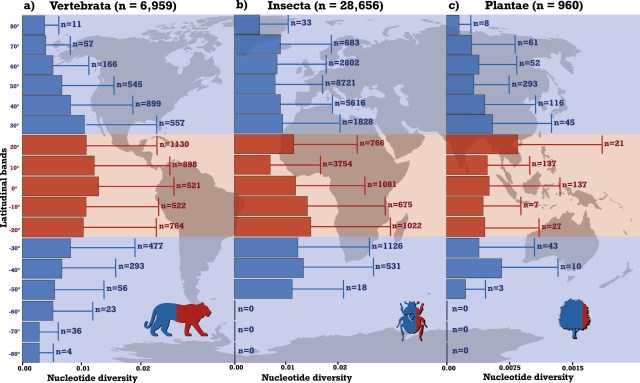
Across all those samples, nucleotide diversity is highest in the tropics, between 30°N and 30°S latitude, with clear patterns of decline towards the poles. The other two statistics, D and R2, were more likely to have values consistent with recent population expansions in species from non-tropical regions — though this test had somewhat variable results across the major taxonomic groups. Natural selection can create genetic patterns similar to population expansion, especially in Tajima’s D, but the authors note that a majority of the variable sites in their aggregate data set are synonymous substitutions, which are much less likely to be under selection.
As the authors note, their results echo a more traditional understanding of latitudinal diversity gradients — there are simply more species in the tropics than in formerly glacier-bound temperate regions. This is perhaps the most intriguing angle on the latitudinal pattern of genetic diversity within species: a classic hypothesis to explain the latitudinal gradient of species diversity is that tropical regions, which have had more stable climates through the Quaternary glacial cycles, offer species more time to adapt to highly specialized niches within their ecological communities. If high-latitude species have had their genetic diversity slashed by the push and pull of glacial advance and retreat, they not only have less time in stable conditions to adapt into specialized ecological roles — they have less diversity to provide raw material for that adaptation.
That’s a pretty impressive link between one of the smallest scales of biological diversity — the single nucleotide substitution — and some of the largest — the species diversity of ecological communities. And the causal agent linking the two is one of the most common molecules on the planet — solid-state H2O, otherwise known as ice.
References
Fonseca EM, TA Pelletier, SK Decker, DJ Parsons, and BC Carstens. 2023. Pleistocene glaciations caused the latitudinal gradient of within-species genetic diversity. Evolution Letters. doi.org/10.1093/evlett/qrad030
Pelletier TA, DJ Parsons, SK Decker, S Crouch, E Franz, J Ohrstrom, and BC Carstens. 2022. phylogatR: Phylogeographic data aggregation and repurposing. Molecular Ecology Resources. 22(8): 2830-2842. doi.org/10.1111/1755-0998.13673
