Today I’m going to write about one of my favorite bioinformatic tools, MultiQC. If you’ve used it, you know why, and if you haven’t, prepare to be amazed.
Many bioinformatic software produce output on a per-sample basis. That is, you may be quality-filtering, trimming, blasting, and mapping your sequence reads to a genome separately for each and every sample. And if you have more than 3 samples (who hasn’t?) going through the output for all samples can become quite tedious and time-consuming.
A single paired-end Illumina MiSeq run can yield 386 separate samples, and bigger genome, transcriptome, and amplicon projects can these days aggregate hundreds or thousands of separate samples.
This is where MultiQC comes in. This tool will first take all the output files from your favorite quality-screening program, and aggregate the results into one simple and pretty report. With FastQC, for example, you normally retrieve a couple of quality plots per sample. MultiQC uses its magic (almost) to compile all samples into combined plots.

Example plot from running FastQC followed by MultiQC. Each line represents one sample.
This way of visualizing your sample output all at once is not only efficient and saves time. It can also easily help you identify outliers, or samples that have various problems, or overall patterns in your data. As an example, in my data I can easily spot which blood samples that contain malaria parasites, by just looking at one single GC content plot aggregated by MultiQC.
However, MultiQC is so much more than that. At the time of writing, it currently supports 38 (!) different tools. Doing RNA-seq analyses? Variant calling? Genome mapping? Bisulfite sequencing? MultiQC compiles the results of your favorite software to give you a quick visual overview of all samples.
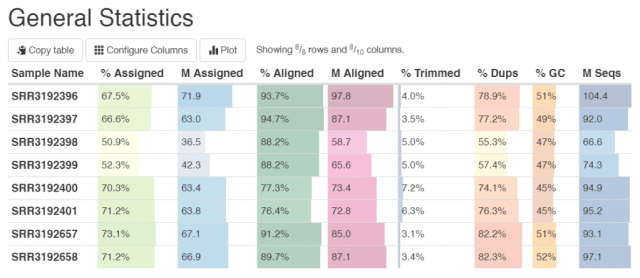
Example output from a mapping software followed by MultiQC.
Example output from the aligner STAR followed by MultiQC.
MultiQC is developed by Phil Ewels, and can be found here. It is ridiculously easy to install with pip. Please remember to cite all tools you use in your work.
Disclaimer: No disclaimer, I just think this is a great tool that deserves more attention. All figures in this post have been shamelessly borrowed from the MultiQC website.
References
Philip Ewels, Måns Magnusson, Sverker Lundin and Max Käller (2016) MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. doi: 10.1093/bioinformatics/btw354

