Recently I saw a graph that I’ve learnt is called the Hype Cycle and is a methodology used in assessment of new technologies and their marketing. What strikes me about it is how well it fits my own research field, paleogenetics or the ancient DNA research.
The Hype Cycle is a graphical tool developed by Gartner, an information technology research and advisory company based in Connecticut. The Hype Cycle depicts five phases of evolution of a new technology, concentrating on the relationship between hype and real adoption of the technology.
Phase I: Innovation Trigger
A potential technology breakthrough kicks things off. Early proof-of-concept stories and media interest trigger significant publicity. Often no usable products exist and commercial viability is unproven. (© Gartner)
It’s not difficult to identify what was the Innovation Trigger in ancient DNA. The origins of the field trace back to mid-1980s when reports of DNA from quagga, an extinct equid, and Egyptian mummies were published.
While the first study, on a 120-year-old quagga (Higuchi et al. 1984), came from the lab of the prominent evolutionary biologist Allan Wilson, the work on Egyptian mummies (Pääbo 1985) was done by then a PhD student, Svante Pääbo, as a secret side-project. Later, Pääbo went to Wilson’s lab and the collaboration of these two men yielded some outstanding studies, regularly occurring in Nature.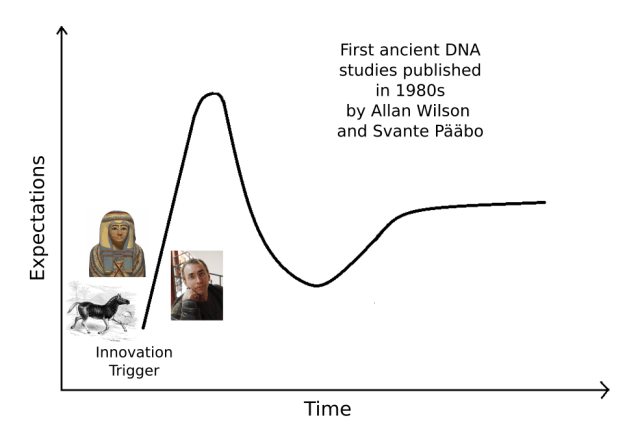
Phase II: Peak of Inflated Expectations
Early publicity produces a number of success stories — often accompanied by scores of failures. Some companies take action; many do not. (© Gartner)
In the following years, the world has seen DNA sequences of the Tasmanian wolf (Thomas et al. 1989), New Zealand moa (Cooper et al. 1992), and the woolly mammoth (Hagelberg et al. 1994) resurrected, but the scientists didn’t restrain to studying animals. Ancient DNA has been also retrieved from plants, e.g. maize (Rollo et al. 1988), and (fanfares) humans (Hagelberg et al. 1989).
Following these amazing achievements, the whole thing got out of hand. Scientists kept publishing more and more sensational results. Especially popular was studying insects in amber, e.g. bees (Cano et al. 1992), a weevil (Cano et al. 1993), and termites (DeSalle et al. 1992). Fossil plants were represented by a Miocene Magnolia species (Golenberg et al. 1994). No sooner did this madness stop, than the first dinosaur DNA was published in Science (Woodward et al. 1994).
Phase III: Trough of Disillusionment
Interest wanes as experiments and implementations fail to deliver. Producers of the technology shake out or fail. Investments continue only if the surviving providers improve their products to the satisfaction of early adopters. (© Gartner)
As we all know, pride goes before a fall, which also applies in this case. In the early 1990s, the young field of ancient DNA believed to have no limits. Everything was possible, as long as nobody tried to replicate those phenomenal studies.

In case of the dinosaur DNA, it was actually much simpler. The original study published in Science didn’t even show a phylogenetic tree, just discussed percentage of sequence difference to other cytochrome b sequences in GenBank. When other scientists (Hedges & Schweitzer 1995) created a phylogenetic tree including all tetrapod sequences in BLAST, the dinosaur didn’t end up together with birds or reptiles, but with a human.
Two important papers published by Tomas Lindahl in Nature (Lindahl 1993a, b) were a rude awakening from many people’s dreams. In a Nature review (Lindahl 1993a), Lindahl explained the process of DNA decay caused by hydrolysis, oxidation, and methylation, once the cell repair mechanisms stop operating after death. In other words, Lindahl pointed out that there is a limit to what can be recovered with ancient DNA and according to his estimate it was more in terms of tens of thousands of years rather than millions.
In the other Nature paper, Lindahl coined the expression “antediluvian DNA” for those hyped contaminated sequenced and called for a more down-to-earth approach to ancient DNA.
“Recent claims of recovery of 100-million-year-old DNA have overshadowed the valuable and important studies on moderately ancient DNA. Rather than proceed spectacularly further and further back in time with anecdotal reports on single samples, using the notoriously contamination-sensitive PCR technique, I suggest that the next goal be a convincing report on the amplification of small DNA fragments, say, 100,000 years old.” (Lindahl 1993b)
Phase IV: Slope of Enlightenment
More instances of how the technology can benefit the enterprise start to crystallize and become more widely understood. Second- and third-generation products appear from technology providers. More enterprises fund pilots; conservative companies remain cautious. (© Gartner)
The “contamination scandals” led several scientists to formulate a set of criteria that were supposed to be obeyed to ensure publication of authentic results. These rules consisted of several steps to control for contamination, e.g. using negative controls and performing analyses in two separate labs.
Scientists became so paranoid that stuff like a paper titled “Ancient DNA: Do It Right or Not at All” (Cooper & Poinar 2000) got published in Science. Meanwhile, other scientists argued that those criteria should be considered more like guidelines and not a checklist, because sometimes fulfilling all criteria doesn’t ensure authenticity, while other times good results couldn’t be published because of unreasonable requirements.
In any case, ancient DNA researchers have learnt their lesson and the coming years brought a lot of good studies, mostly on short mitochondrial DNA fragments. Even today, I use those early studies when lecturing about ancient DNA, because they represent a very simple yet elegant way of testing phylogeographic and demographic models.
Those were also the times when high-throughput sequencing was born and progressing by leaps and bounds. And just as genetics was driven by the Human Genome Project, a lot of development in ancient DNA was thanks to the ambitious Neanderthal Genome Project, led by the same man who was at the very beginning of the field, Svante Pääbo.
Phase V: Plateau of Productivity
Mainstream adoption starts to take off. Criteria for assessing provider viability are more clearly defined. The technology’s broad market applicability and relevance are clearly paying off. (© Gartner)
I dare to say that ancient DNA has reached the Plateau of Productivity. Scientists are testing the limits of what is possible, but this time within sensible limits. After the sensational plans to sequence anything that has ever lived and the subsequent disappointment that we wouldn’t be able to get over the border of 100,000 years, recent work is pushing limits in the right direction again. Currently, we are at 700,000 years back to the past (Orlando et al. 2013).
Thanks to new methods that are able to recover very short DNA fragments, methods to treat the damaged DNA, knowledge of where to look for the right samples, and ever more efficient sequencing technologies, we are living the times of population genetic studies on ancient populations, high-coverage genomes of extinct animals, understanding the history of our own species, and we are again approaching the 1-million border. This time for real.
 References
References
Cano RJ, Poinar H, Poinar Jr. GO. 1992. Isolation and partial characterisation of DNA from the bee Proplebeia dominicana (Apidae: Hymenoptera) in 25–40 million year old amber. Med. Sci. Res. 20, 249–251.
Cano RJ, Poinar HN, Pieniazek N J, Acra A, Poinar Jr. GO. 1993. Amplification and sequencing of DNA from a 120–135 million-year-old weevil. Nature 363, 536–538. doi: 10.1038/363536a0
Cooper A, Mourer-Chauviré C, Chambers GK, von Haeseler A, Wilson AC, Pääbo S. 1992. Independent origins of New Zealand moas and kiwis. Proc. Natl Acad. Sci. USA 89, 8741–8744. doi: 10.1073/pnas.89.18.8741
Cooper A, POINAR HN. 2000. Ancient DNA: Do It Right or Not at All. Science 289(5482), 1139. doi: 10.1126/science.289.5482.1139b
Golenberg EM, Giannasi DE, Clegg MT, Smiley CJ, Durbin M, Henderson D, Zurawski G. 1990. Chloroplast DNA sequence from a Miocene magnolia species. Nature 344, 656–658. doi: 10.1038/344656a0
Hagelberg E, Sykes B, Hedges R. 1989. Ancient bone DNA amplified. Nature 342, 485. doi: 10.1038/342485a0
Hagelberg E, Thomas MG, Cook CE, Sher AV, Baryshnikov GF, Lister AM. 1994. DNA from ancient mammoth bones. Nature 370, 333–334. doi: 10.1038/370333b0
Hedges SB, Schweitzer MH. 1995. Detecting dinosaur DNA. Science 5214, 1191-1192. doi: 10.1126/science.7761839
Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. 1984. DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282–284. doi: 10.1038/312282a0
Lindahl T. 1993a. Instability and decay of the primary structure of DNA. Nature 362(6422): 709–15. doi: 10.1038/362709a0
Lindahl T. 1993b. Recovery of antediluvian DNA. Nature 365: 700. doi:10.1038/365700a0
Orlando et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78. doi: 10.1038/nature12323
Pääbo S. 1985. Molecular cloning of ancient Egyptian mummy DNA. Nature 314, 644–645. doi: 10.1038/314644a0
Rollo F, Amici A, Salvi R, Garbuglia A. 1988. Short but faithful pieces of ancient DNA. Nature 335, 774. doi: 10.1038/335774a0
Thomas RH, Schaffner W, Wilson AC, Pääbo S. 1989. DNA phylogeny of the ancient marsupial wolf. Nature 340, 465–467. doi: 10.1038/340465a0
Woodward SR, Weyand NJ, Bunnell M. 1994. DNA sequence from Cretaceous period bone fragments. Science 265,1229. doi: 10.1126/science.7973705



