 How did we get where we are? Genetically speaking, that is. A few posts ago, that whole genotype-phenotype question was discussed, how do genomes make plants and animals (and don’t forget the microbes!) look and act how they do. Another broader question linked to this topic involves trying to understand how genomes have evolved over time, clearly no small task.
How did we get where we are? Genetically speaking, that is. A few posts ago, that whole genotype-phenotype question was discussed, how do genomes make plants and animals (and don’t forget the microbes!) look and act how they do. Another broader question linked to this topic involves trying to understand how genomes have evolved over time, clearly no small task.
 It’s pretty amazing that we are at a point where we can attempt to unravel that puzzle. It appears that almost daily, complete genome sequences are more and more affordable (and of better quality). In a cool study published online in PNAS this week, Kapusta and colleagues looked at genomes from mammal and bird species and revealed that in both groups, genome expansion (aka when the genome gets larger, in this case due to transposable element (TE) sequences) is offset by large segmental deletions (genomes getting smaller). Basically, demonstrating that these genomes have fluctuated in size a whole lot more than we realized. As a side note, ‘amniotes’ refers to a group of animals (including us) as well as reptiles, birds, and other mammals, just in case you were wondering.
It’s pretty amazing that we are at a point where we can attempt to unravel that puzzle. It appears that almost daily, complete genome sequences are more and more affordable (and of better quality). In a cool study published online in PNAS this week, Kapusta and colleagues looked at genomes from mammal and bird species and revealed that in both groups, genome expansion (aka when the genome gets larger, in this case due to transposable element (TE) sequences) is offset by large segmental deletions (genomes getting smaller). Basically, demonstrating that these genomes have fluctuated in size a whole lot more than we realized. As a side note, ‘amniotes’ refers to a group of animals (including us) as well as reptiles, birds, and other mammals, just in case you were wondering.
“The results are consistent with an ‘accordion’ model of genome size equilibrium, whereby DNA gains are balanced by DNA loss, primarily through large-size deletions.”
 Transposable element (TE) sequences are kind of amazing. They are essentially sections of DNA that can literally move within a genome, leading to duplications of regions of DNA and ultimately changing the size of a genome. They were first identified in maize by Barbara McClintock in the 1950’s, and make up a HUGE portion of that plant’s genome (~85%). Although it’s been shown that TEs have played a role in the evolution of genomes in both mammals and birds, how it hasn’t led to huge changes in genome size was (until recently) confounding.
Transposable element (TE) sequences are kind of amazing. They are essentially sections of DNA that can literally move within a genome, leading to duplications of regions of DNA and ultimately changing the size of a genome. They were first identified in maize by Barbara McClintock in the 1950’s, and make up a HUGE portion of that plant’s genome (~85%). Although it’s been shown that TEs have played a role in the evolution of genomes in both mammals and birds, how it hasn’t led to huge changes in genome size was (until recently) confounding.
 Every organism has a genome, but they can be pretty dang different in size and architecture (how it’s organized). The authors highlight that previous studies often compared species of very diverse genome sizes, such as barley (with a 4,900 Mb genome) with pufferfish (with a 400 Mb genome), and found that larger genome species have a larger proportion of their genome dedicated to TE sequences. While, it has been shown that it’s both natural selection and adaptation that are key in determining genome size, understanding what is responsible for shifts in genome size on a finer scale is not clear.
Every organism has a genome, but they can be pretty dang different in size and architecture (how it’s organized). The authors highlight that previous studies often compared species of very diverse genome sizes, such as barley (with a 4,900 Mb genome) with pufferfish (with a 400 Mb genome), and found that larger genome species have a larger proportion of their genome dedicated to TE sequences. While, it has been shown that it’s both natural selection and adaptation that are key in determining genome size, understanding what is responsible for shifts in genome size on a finer scale is not clear.
Interestingly, Kapusta et al., decided to tackle the analysis of a whole different beast (technically a bunch of them, I suppose), by analyzing groups with relatively low levels of interspecific genome size variation. The authors assessed how much size variation has occurred over the past 100 million years among the genomes of 10 mammal species and 24 species of birds.
“Paradoxically, birds and bats have more compact genomes relative to their flightless relatives but exhibit more dynamic gain and loss of DNA.”
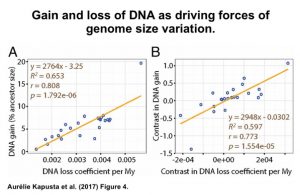
 In order to compare genome sizes over time, Kapusta et al., created a model to determine genome size change by comparing the genomes of current organisms to that of their last common ancestors. Their results reveal that among mammal and bird genomes there has been plenty of gains as well as losses, which has led to these genomes reaching a state of equilibrium. The authors refer to this expansion and contraction of the genome as an “accordion” model of genome size evolution.
In order to compare genome sizes over time, Kapusta et al., created a model to determine genome size change by comparing the genomes of current organisms to that of their last common ancestors. Their results reveal that among mammal and bird genomes there has been plenty of gains as well as losses, which has led to these genomes reaching a state of equilibrium. The authors refer to this expansion and contraction of the genome as an “accordion” model of genome size evolution.
“In other words, the genomes of flightless birds are less dynamic overall than those of flying species.”
Interestingly, they highlight that both bats and the flying birds appear to have more streamlined genomes. The authors suggest that the high energy requirement of flight might be involved in why these genomes are more constrained in size.
As techniques to tease apart the intricate mechanisms driving changes in genomes develop, our understanding of the forces that influence genomes also evolves. And that, I think, is music to everyone’s ears.
References:
Kapusta, A., Suh, A., Feschotte, C., 2017. Dynamics of genome size evolution in birds and mammals. PNAS. Published online February 8, 2017. DOI: 10.1073/pnas.1616702114
Bennetzen, J.L., Ma, J. and Devos, K.M., 2005. Mechanisms of recent genome size variation in flowering plants. Annals of botany, 95(1), pp.127-132. DOI: https://doi.org/10.1093/aob/mci008
Kazazian, H.H., 2004. Mobile elements: drivers of genome evolution. science, 303(5664), pp.1626-1632. DOI: 10.1126/science.1089670
