
One of the grand patterns across the diversity of flowering plants is that major groups of species are deeply united by shared chemistry, especially “secondary” biochemical products that don’t directly contribute to processes like photosynthesis, growth, and reproduction. Secondary compounds often have defensive function, and they’ve long been recognized as the key to the evolutionary history of plant-herbivore interactions. According to a nifty new study, biochemistry’s deep linkage to plants’ evolution may also make it the most useful index of a plant community’s functional diversity.
The idea that flowering plants’ chemistry reflects deep historical relationships among species is an old one. The earliest articulation I know is an analysis by Helen C. de S. Abbott published in The American Naturalist back in 1887, which attempts to link increasing biochemical complexity — such as could be assayed in the late nineteenth century — to evolutionary “higher forms”. Abbott’s theorizing was limited, in part, by that nineteenth century view of “higher” versus “lower” forms, but she was on to something. Almost eight decades later, Paul Ehrlich and Peter Raven noted that shared anti-herbivore defenses in major groups of plants were mirrored by the feeding preferences of major groups of butterflies, and concluded this reflected the coevolution of a multi-million-year plant-herbivore arms race.
Ehrlich and Raven published back in 1964, but while we’ve substantially revised and refined the high-level taxonomy of flowering plants since then, the patterns that they, and Abbott, noticed have held up. I come back to the showiest examples multiple times in a semester of Flowering Plant Systematics: the cardiac glycosides of the milkweed family, the diverse and often deadly alkaloids of the nightshades, or the phototoxic furanocoumarins of the umbellifers. It’s a textbook axiom that plants’ biochemistry reflects their evolutionary history.
This is the jumping-off point for the new study. Ecologists George Furey and David Tilman set out to test how well plants’ chemistry reflects the ecology of plant communities. Community ecology has long been interested in links between morphological and functional diversity — the variety of forms of plants and their contributions to local nutrient cycles — and the overall productivity of a community — literally, how much plant mass can it generate?
Since before Charles Darwin, we’ve understood that a plot of ground hosting a greater diversity of plant species can (usually) generate more biomass than an otherwise identical plot hosting fewer species. What specific features of a diverse plant community make this possible, though, isn’t entirely clear. Finding features of the plants in a community that reliably predict its overall productivity would suggest that the diversity of those features represented by the plant species in the community is ultimately responsible for its productivity.
Furey and Tilman started with data from a classic ecological experiment comparing productivity by experimental plant communities composed of anywhere from one to 15 species from North American prairies. That experiment found that communities with more species were more productive; Furey and Tilman assembled data on the morphology, metabolic performance, and chemistry of the species in the experimental communities, and then set out to identify which set of characteristics most usefully explained differences in productivity. They broadly categorized plants’ features as (1) morphological traits like leaf shape or plant height; (2) metabolic traits like photosynthetic rate; and (3) chemical traits that were all tissue content of individual elements: aluminum, boron, carbon, nitrogen, et cetera.
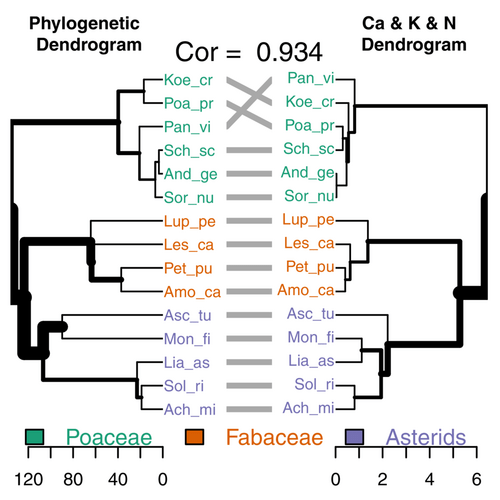
Furey and Tilman tested how well the species’ phylogenetic history matched their differences in all possible sets of three traits — more than 11,000 combinations of three from the total set of 43 traits. From that very long list of possibilities, the best-fit trait set was all chemical: the tissue content of calcium, potassium, and nitrogen had a cophenetic correlation of more than 0.9 with the phylogeny of the 15 species. The resulting “tanglegram” is the paper’s Figure 1, above. You can see how well chemical differences align with the phylogeny of the species, which include five in the Asterid clade, four members of the nitrogen-fixing Fabaceae, and six grasses (the Poaceae). Chemical traits were 82% of the traits in the 15 three-trait sets that best matched the phylogeny; just 2% were metabolic traits, and the rest were morphological.
The authors then compare representation of the three chemical trait-based clusters (which are also major taxonomic groups) to overall productivity in the experimental communities. While across all the possible plant communities, there’s a clear signal of increasing biomass production with the addition of more species, it emerges that this is driven by the diversity of tissue-chemistry clusters, rather than simple species counts. A community of four species from a single cluster would produce less total biomass than a community of four species that had members of two clusters, which was less productive than a community of four species including members of all three clusters.

That’s a neat result — but it’s also very clearly driven by the fact that the chemical traits used to cluster the 15 species align almost perfectly with their phylogeny. It maybe sounds less impressive if I say that Furey and Tilman found a community of two grasses, a legume, and an aster is more productive than a community of four legumes. But the goal here is to get closer to understanding why phylogenetically diverse communities are more productive.
As Furey and Tilman note in their conclusion, most of the focus in community ecology has been, up to now, on more obviously functional phenotypes — morphological diversity, and metabolic diversity. They’ve shown that the chemical content of plants’ tissues is more closely related to their phylogenetic history than traits in those categories (at least the ones they examine). Chemical composition thereby ends up better predicting a community’s total productivity. It may be, Furey and Tilman suggest, that these biochemical differences reflect ancient niche differentiation between the three big clades represented in the experimental communities.
Those biochemical differences probably mean that grasses, legumes, and asterids need somewhat different suites of chemical nutrients from the soil, and maybe they’re less likely to compete when planted together as a result. Or, if the biochemical diversity is diversity of secondary chemistry, it may let members of the three clades maintain a kind of collective defense — a plot full of legumes may be a more tempting target for legume-specialist herbivores than a plot of legumes mixed with other taxa. The original community composition experiment did not, as far as I can tell from its methods, exclude or otherwise control insect herbivory. And, somewhat surprisingly to me, the new paper’s only discussion of defensive chemistry is in passing remarks.
All in all, an interesting new result — I do think it gets us closer to understanding how biodiversity relates to community productivity, but it’s far from the last word on the question.
References
Abbott HC de S. 1887. Comparative chemistry of higher and lower plants. The American Naturalist. 21(8): 719-730. doi.org/10.1086/274542
Ehrlich PR and PH Raven. 1964. Butterflies and plants: a study in coevolution. Evolution, 18(4): 586–608. doi.org/10.1111/j.1558-5646.1964.tb01674.x
Furey GN and D Tilman. 2023. Plant chemical traits define functional and phylogenetic axes of plant biodiversity. Ecology Letters. doi.org/10.1111/ele.14262
Tilman D, J Knops, D Wedin, P Reich, M Ritchie and E Siemann. 1997 The influence of functional diversity and composition on ecosystem processes. Science. 277: 1300–1302. doi.org/10.1126/science.277.5330.1300
